FNDC4 acts on Orphan GPR116 to enhance insulin sensitivity
Abstract
Orphan GPR116 mediates the insulin sensitizing effects of the hepatokine FNDC4 in adipose tissue
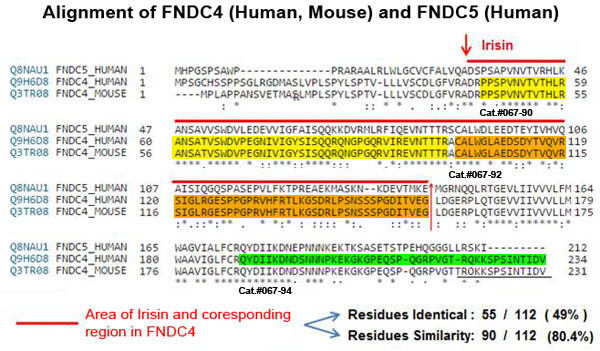
*The authors validate the quantified levels of sFNDC4 by using Phoenix’s FNDC4 (47-101) (Human) / FNDC4 (43-97) (Mouse) – EIA Kit (catalog #EK-067-90) and found zero cross-reactivity to irisin/FNDC5
Georgiadi A, Lopez-Salazar V, Merahbi RE-, et al. Orphan GPR116 mediates the insulin sensitizing effects of the hepatokine FNDC4 in adipose tissue. Nat Commun. 2021;12(1):2999.
Supplementary Information: https://static-content.springer.com/esm/art%3A10.1038%2Fs41467-021-22579-1/MediaObjects/41467_2021_22579_MOESM1_ESM.pdf
FNDC4 acts as an anti-inflammatory factor on macrophages and improves colitis in mice.
FNDC4 is a secreted factor sharing high homology with the exercise-associated myokine irisin (FNDC5). Here we report that Fndc4 is robustly upregulated in several mouse models of inflammation as well as in human inflammatory conditions. Specifically, FNDC4 levels are increased locally at inflamed sites of the intestine of inflammatory bowel disease patients. Interestingly, administration of recombinant FNDC4 in the mouse model of induced colitis markedly reduces disease severity compared with mice injected with a control protein. Conversely, mice lacking Fndc4 develop more severe colitis. Analysis of binding of FNDC4 to different immune cell types reveals strong and specific binding to macrophages and monocytes.FNDC4 treatment of bone marrow-derived macrophages in vitro results in reduced phagocytosis, increased cell survival and reduced proinflammatory chemokine expression. Hence, treatment with FNDC4 results in a state of dampened macrophage activity, while enhancing their survival. Thus, we have characterized FNDC4 as a factor with direct therapeutic potential in inflammatory bowel disease and possibly other inflammatory diseases.
Bosma M1, Gerling M2, Pasto J1 et al., Nat Commun. 2016 Apr 12;7:11314. doi: 10.1038/ncomms11314.
Discovery and fine mapping of serum protein loci through transethnic meta-analysis
Many disorders are associated with altered serum protein concentrations, including malnutrition, cancer, and cardiovascular, kidney, and inflammatory diseases. Although these protein concentrations are highly heritable, relatively little is known about their underlying genetic determinants. Through transethnic meta-analysis of European-ancestry and Japanese genome-wide association studies, we identified six loci at genome-wide significance (p < 5 × 10(-8)) for serum albumin (HPN-SCN1B, GCKR-FNDC4, SERPINF2-WDR81, TNFRSF11A-ZCCHC2, FRMD5-WDR76, and RPS11-FCGRT, in up to 53,190 European-ancestry and 9,380 Japanese individuals) and three loci for total protein (TNFRS13B, 6q21.3, and ELL2, in up to 25,539 European-ancestry and 10,168 Japanese individuals). We observed little evidence of heterogeneity in allelic effects at these loci between groups of European and Japanese ancestry but obtained substantial improvements in the resolution of fine mapping of potential causal variants by leveraging transethnic differences in the distribution of linkage disequilibrium. We demonstrated a functional role for the most strongly associated serum albumin locus, HPN, for which Hpn knockout mice manifest low plasma albumin concentrations. Other loci associated with serum albumin harbor genes related to ribosome function, protein translation, and proteasomal degradation, whereas those associated with serum total protein include genes related to immune function. Our results highlight the advantages of transethnic meta-analysis for the discovery and fine mapping of complex trait loci and have provided initial insights into the underlying genetic architecture of serum protein concentrations and their association with human disease.
Rizkalla Salwa W, Edi Prifti, Aurélie Cotillard et al., et al., Am J Hum Genet. 2012 Oct 5;91(4):744-53. doi: 10.1016/j.ajhg.2012.08.021. Epub 2012 Sep 27.
Genome-wide methylation profiling of schizophrenia.
Schizophrenia is one of the major psychiatric disorders. It is a disorder of complex inheritance, involving both heritable and environmental factors. DNA methylation is an inheritable epigenetic modification that stably alters gene expression. We reasoned that genetic modifications that are a result of environmental stimuli could also make a contribution. We have performed 26 high-resolution genome-wide methylation array analyses to determine the methylation status of 27,627 CpG islands and compared the data between patients and healthy controls. Methylation profiles of DNAs were analyzed in six pools: 220 schizophrenia patients; 220 age-matched healthy controls; 110 female schizophrenia patients; 110 age-matched healthy females; 110 male schizophrenia patients; 110 age-matched healthy males. We also investigated the methylation status of 20 individual patient DNA samples (eight females and 12 males. We found significant differences in the methylation profile between schizophrenia and control DNA pools. We found new candidate genes that principally participate in apoptosis, synaptic transmission and nervous system development (GABRA2, LIN7B, CASP3). Methylation profiles differed between the genders. In females, the most important genes participate in apoptosis and synaptic transmission (XIAP, GABRD, OXT, KRT7), whereas in the males, the implicated genes in the molecular pathology of the disease were DHX37, MAP2K2,FNDC4 and GIPC1. Data from the individual methylation analyses confirmed, the gender-specific pools results. Our data revealed major differences in methylation profiles between schizophrenia patients and controls and between male and female patients. The dysregulated activity of the candidate genes could play a role in schizophrenia pathogenesis.
Rukova B, Staneva R, Hadjidekova S, et al., Balkan J Med Genet. 2015 Apr 10;17(2):15-23. doi: 10.2478/bjmg-2014-0070. eCollection 2014.
Teufel A1, Malik N, Mukhopadhyay M et al., Gene. 2002 Sep 4;297(1-2):79-83.
Schematics

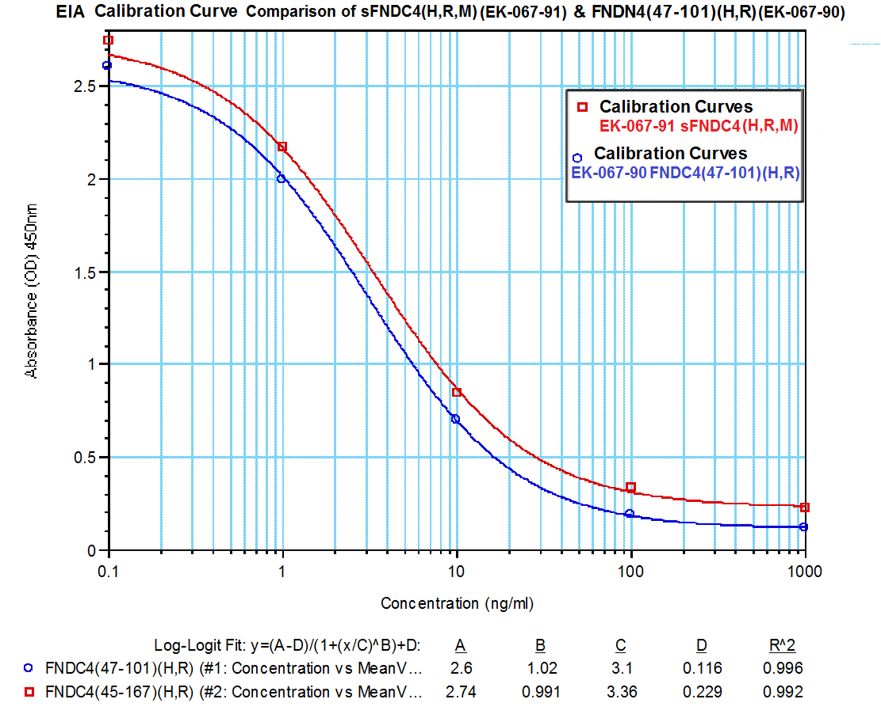
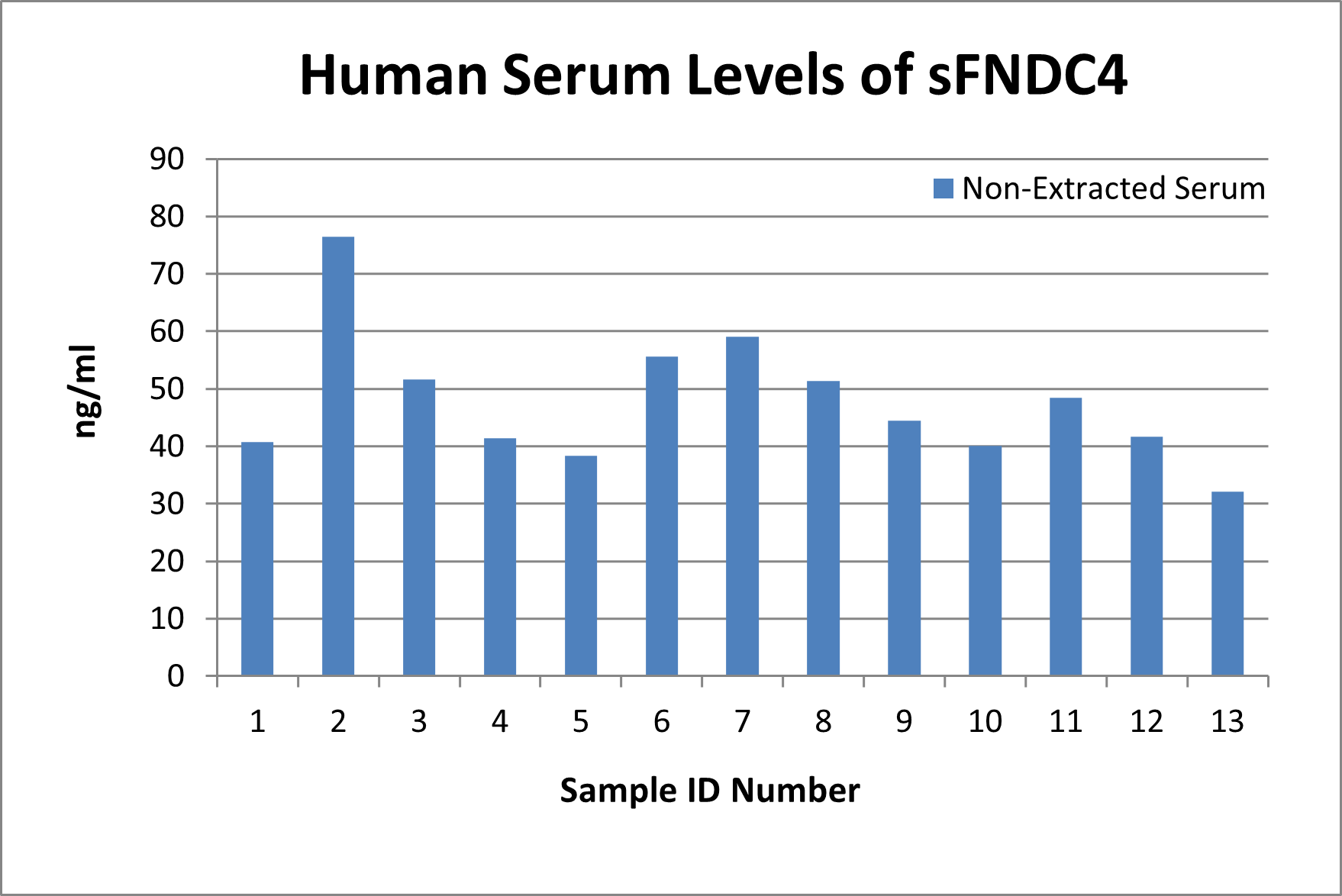
In-house measured levels of sFNDC4 using Catalog No. EK-067-91
Human Serum levels: 47.78 ± 3.06 ng/ml (n=13)
